Phantom QA
Human QA
- fMRI
- Motion related metrics
- AFNI euclidean (mm)
- FD and DVARS
- niftitools’ QA (niftiqa)
- fBIRN QA for EPI images: mirror of fBIRN QA metrics explanations.
- Motion related metrics
- sMRI
- dMRI
Robust pipelines:
- ini default config files (resting-state, task)
- sMRI (including snapshots for each step)
- Skull strip (ROBEX / BEaST/FSL BET/AFNI 3dSkullStrip/FreeSurfer stage 1-5)
- non-local means filtering (optional)
- Robust MRI registration (FreeSurfer)
- FSL 3-stages registration
- dof-9
- dof-12
- FNIRT
- FAST segmentation
- preparing for BBR registration
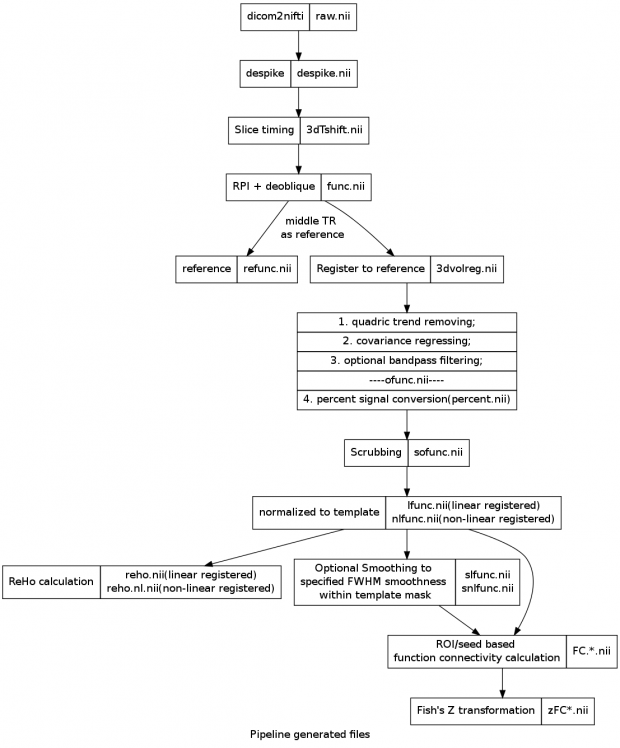
- dicom to nifti, automatically, extract information from header
- slice timing correction to 1st slice
- motion metrics scrubbing/censoring (FD, DVARS, 3dvolreg) and regressors generation (6-motion, FD, DVARS, Friston-24)
- CSF/WM nuiances PCA signals extration (default using first 6 PCs)
- BBR to anat, then normalized to template (one step to minimize distortion of fMRI signals)
- Optional smoothing
- ReHo calculation
- FC calculation and Fish’s Z transformation
- dMRI
Linux misc:
- Ubuntu recommanded since there is a NeuroDebian project, which will make neuroimaging life much easier
- Viewing windows in a remote shell of linux
- Easy way: Install MobaXterm
- Advanced way: install PuTTy and Cygwin which I prefer.
- With Virtualbox and PuTTy/MobaXterm, we can work seamlessly with a local linux server including viewing results of FreeSurfer/Caret. Just remember map 22 port from virtual-linux to windows.
QUEST misc
- msubi, get a shell of a computing node
- msubq, submit a command line to run in a computing node
- qjobs, get my jobs’ status and working dir …
- qnodeslive, get current working node to debug programs
NUNDA misc
I wrote a command line version of XNAT database operation interface based on curl with XNAT API and pyxnat. I am trying to make using NUNDA easier.
- xnatlogin
- xnatview.py -p project -q subject -s session
- xnatget.py projID subjID sessID
- xnatsess.py -r sessID
- xnatput.py
NUNDA resources levels in a nutshell:
- - projects - - subjects - - experiments - - scans - - reconstructions - - assessors
Always keep Backup in mind:
- By duplicati (Basic, easy to use): http://www.duplicati.com/
- By duplicity (Advanced, easy to make script):