This is the general user guide to do online data analysis using Robust pipelines.
To be noted, the “online” does not mean real-time fMRI, but using web based interface which even allow submitting jobs 6pm on the train with smart phones, and check results in next day. However, though this is for offline analysis, the QA can be done quickly/automatically to decide if it is necessary to re-scan the subject before he/she left.
Outline
0. Basic knowledge about NUNDA/XNAT, which can be easily grasped after playing/poking a while with NUNDA.
0-a. after login
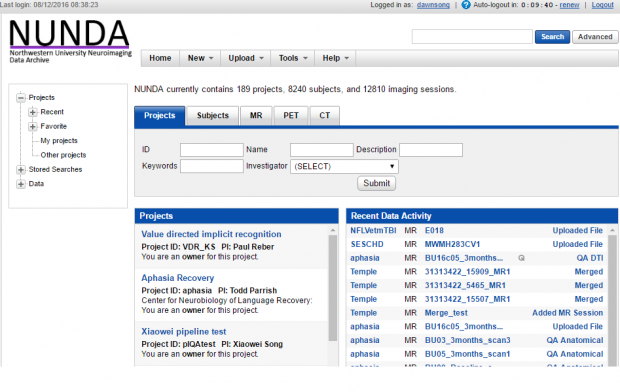
0-b. NUNDA resources levels in a nutshell:
- - projects - - subjects - - experiments - - scans - - reconstructions - - assessors
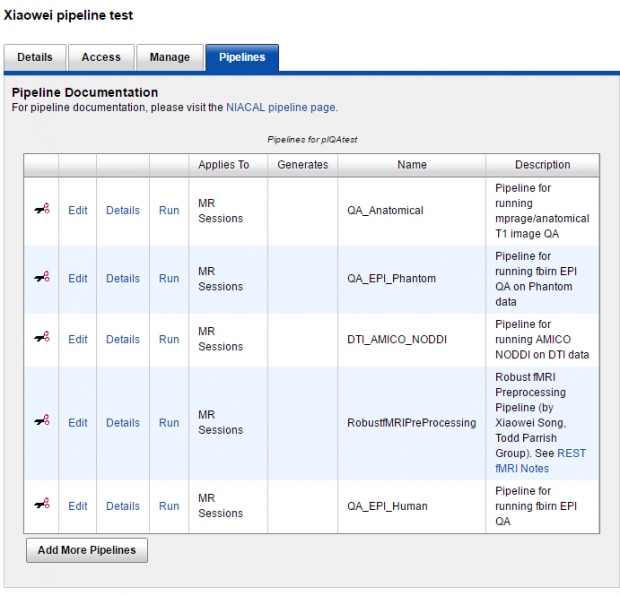
0-c. "Adding" a pipeline to a project in NUNDA,
or "edit" default parameters for one pipeline,
or launch ("run") a pipeline in project level for all subjects' all matched scans.
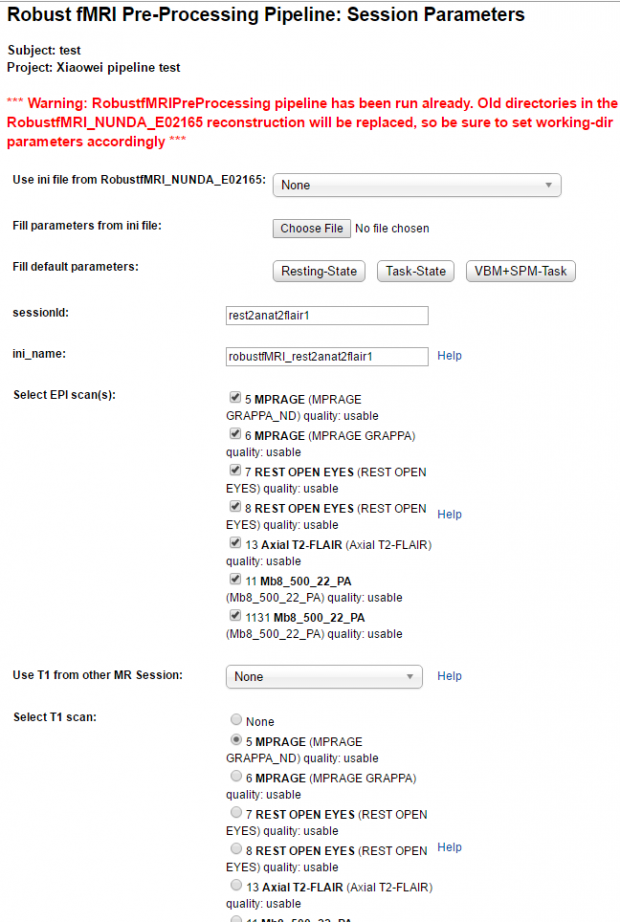
0-d. launch Robust pipeline (the other similar) in one session to select anat/func DICOMs/scans.
1. Gathering the data (phantom; Anatomical, Functional, DWI of human brain).
1-a. MRI was automatically setup to upload data (Contact Dr. Parrish about how to setup).
1-b. User need upload data manually to NUNDA.
2. QA, if automated, usually can be done quickly to allow decision of re-scan or not. Except multi-band and DWI data, which may need 1~2 hours besides waiting time in Quest queue.
5. DWI preprocessing
5-a. Camino based Robust tensor estimation, plus normal PCA based FA/MD/… analysis.
5-b. NODDI/AMICO based tensor analysis.
6. 1st level analysis by SPM/FEAT with filename mapping, which should do exactly same job if the batch can run locally for one subject, but here can be run simultaneously for 1000 subjects, while time can be as short as 1 subject, thanks to the power of Quest HPC.
7. Robust helper functions
8. 2nd/group level statistics
9. downloading results in script.
FAQ with brief answer:
1. Why another pipeline or another set of pipelines?
2. Why using web interface?
3. Why calling Robust?
4. Why using HPC?
5. Why some PDF font not readable?
6. Is it true almost all applications are run-able including OpenGL or graphical interfaced programs?
7. What’s the difference between rlesion.msk+tpl.nii and lesion.msk2tpl.nii ?
rlesion.msk+tpl.nii is interpolated by tri-linear if using FSL, B-spline (degree of 5) if using SPM/DARTEL.
lesion.msk2tpl.nii is interpolated by using Nearest-Neighbor method no matter FSL or SPM/DARTEL was used.
The difference between interpolation means rlesion.msk+tpl might look smoother in edges than lesion.msk2tpl, but it can also have rings artefact, unlike NN, which does not create new “intensities” as smooth interpolation does.
For example, for mask image which only have 0/1 as values, after interpolation other than NN, it will have values from 0 to 1, instead of binary.
8. my Works/Changes list.
References:
- Xiaowei Song, Xue Wang, Kate Alpert, Yufen Chen, Lejian Huang, Lei Wang, Todd Parrish. Rapid automatic comprehensive quality assurance metrics evaluation for Neuroimaging studies. (Organization for Human Brain Mapping Annual Meeting, 2015)
- Xiaowei Song, Xue Wang, Kate Alpert, Lei Wang, Todd Parrish. Robust web-based parallel-optimized minimal pre-processing and analyzing pipeline for fMRI big data. (Organization for Human Brain Mapping Annual Meeting, 2015)